Harminv
From AbInitio
| Revision as of 22:35, 25 February 2015 (edit) Stevenj (Talk | contribs) (→Download) ← Previous diff |
Revision as of 22:35, 25 February 2015 (edit) Stevenj (Talk | contribs) (→Download) Next diff → |
||
| Line 31: | Line 31: | ||
| A Python interface to Harminv was developed by Aaron O'Leary: [https://github.com/aaren/harminv pharminv]. | A Python interface to Harminv was developed by Aaron O'Leary: [https://github.com/aaren/harminv pharminv]. | ||
| - | Please file bug reports or feature requests as [harminv Github issues](https://github.com/stevengj/harminv/issues). | + | Please file bug reports or feature requests as [https://github.com/stevengj/harminv/issues harminv Github issues]. |
| = Documentation = | = Documentation = | ||
Revision as of 22:35, 25 February 2015
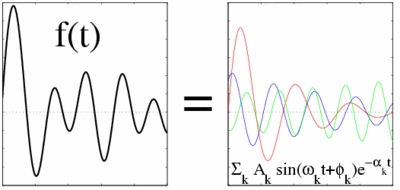
Harminv is a free program (and accompanying library) to solve the problem of harmonic inversion — given a discrete-time, finite-length signal that consists of a sum of finitely-many sinusoids (possibly exponentially decaying) in a given bandwidth, it determines the frequencies, decay constants, amplitudes, and phases of those sinusoids.
It can, in principle, provide much better accuracy than straightforwardly extracting FFT peaks, essentially because it assumes a specific form for the signal. (Fourier transforms, in contrast, attempt to represent any data as a sum of sinusoidal components, and are thus limited by the uncertainty principle.) It is also often more robust than directly least-squares fitting the data (which can have problematic convergence), since it re-expresses the problem in terms of simply finding the eigenvalues of a small matrix.
Harminv uses a low-storage "filter diagonalization method" (FDM) for finding the sinusoids near a given frequency interval, based on the algorithm described in (see also below):
- V. A. Mandelshtam and H. S. Taylor, "Harmonic inversion of time signals," J. Chem. Phys. 107 (17), 6756-6769 (1997). Erratum, ibid. 109 (10), 4128 (1998).
This kind of spectral analysis has wide applications in many areas of physics and engineering, as well as other fields. For example, it could be used to extract the vibrational or "eigen" modes of a system from its response to some stimulus, and also their rates of decay in dissipative systems. FDM has been applied to analyze, e.g., NMR experimental data. It is especially appropriate for analyzing numerical simulations, e.g. of quantum mechanics or classical electromagnetism. In general, it is useful when you know on physical grounds that your system consists of a small number of decaying & oscillating modes in the bandwidth of interest, plus a limited amount of noise, and is not appropriate to analyze completely arbitrary waveforms.
Harminv was developed by Steven G. Johnson (stevenj@alum.mit.edu), and is free software that should install under any Unix-like operating system (e.g. GNU/Linux).
Download
The latest version is Harminv 1.3.1, which can be downloaded in source-code form at:
What's new in each version is described in the Harminv release notes. Harminv is distributed under the GNU GPL and comes with NO WARRANTY (see the license for more details). Development sources can be found on GitHub:
It would be courteous of you to cite Harminv and its author in any publication for which you find it useful, in addition to citing a Mandelshtam reference (either the one above or the review article below).
To install, please see our installation instructions. The main requirement is that you first install the BLAS and LAPACK libraries.
If you use Debian GNU/Linux, you can use the Debian package of harminv (currently in Debian/unstable), packaged by Loic Le Guyader.
A Python interface to Harminv was developed by Aaron O'Leary: pharminv.
Please file bug reports or feature requests as harminv Github issues.
Documentation
Most users will employ Harminv via the standalone harminv program, which is described by the harminv man page. For instructions on calling the libharminv library directly from your own C/C++ code, see the README file included with Harminv.
Essentially, the FDM algorithm works by considering the time-series to be the autocorrelation function of a fictitious dynamical system, such that the problem of finding the frequencies and decay constants is re-expressed as the problem of finding the eigenvalues of the complex-symmetric time-evolution operator of this system. The key point is that, if you are only interested in frequencies within a known band-limited region, the matrix elements of this operator can be expressed purely in terms of Fourier transforms (or, really, z transforms) of your time-series. Then, one can simply diagonalize a small matrix (size proportional to the bandwidth and the number of frequencies) to find the desired result.
In general, for M data points and J frequencies, the time required is O(M J) + O(J3). The main point of the algorithm is not really speed, however, but rather the effective solution of a typically ill-conditioned fitting problem. (Even closely-spaced frequencies, large numbers of modes, and/or weak decay rates can often be resolved reliably by FDM, whereas a straightforward fit of the data or its spectrum is problematic in such cases.)
Some additional references on this method are:
- Michael R. Wall and Daniel Neuhauser, "Extraction, through filter-diagonalization, of general quantum eigenvalues or classical normal mode frequencies from a small number of residues or a short-time segment of a signal. I. Theory and application to a quantum-dynamics model," J. Chem. Phys. 102 (20), 8011-8022 (1995). (This is a precursor of the Mandelshtam & Taylor method.)
- V. A. Mandelshtam and H. S. Taylor, "Harmonic inversion of time signals," J. Chem. Phys. 107 (17), 6756-6769 (1997). Erratum, ibid. 109 (10), 4128 (1998).
- H. Hu, Q. N. Van, V. A. Mandelshtam, and A. J. Shaka, "Reference deconvolution, phase correction, and line listing of NMR spectra by the 1D filter diagonalization method," J. Magnetic Resonance 134, 76-87 (1998).
- Rongqing Chen and Hua Guo, "Efficient calculation of matrix elements in low storage filter diagonalization," J. Chem. Phys. 111 (2), 464-471 (1999).
- V. A. Mandelshtam, "FDM: the filter diagonalization method for data processing in NMR experiments," Progress in Nuclear Magnetic Resonance Spectroscopy 38, 159-196 (2001). (Review article.)
Newer twists that we have not yet implemented include:
- J. Chen and V. A. Mandelshtam, "Multiscale filter diagonalization method for spectral analysis of noisy data with nonlocalized features," J. Chem. Phys. 112 (10), 4429-4437 (2000).
- V. A. Mandelshtam, "On harmonic inversion of cross-correlation functions by the filter diagonalization method," J. Theoretical and Computational Chemistry 2 (4), 497-505 (2003).
...as well as other variants, such as multidimensional FDM, described in the 2001 review article above.